[ad_1]
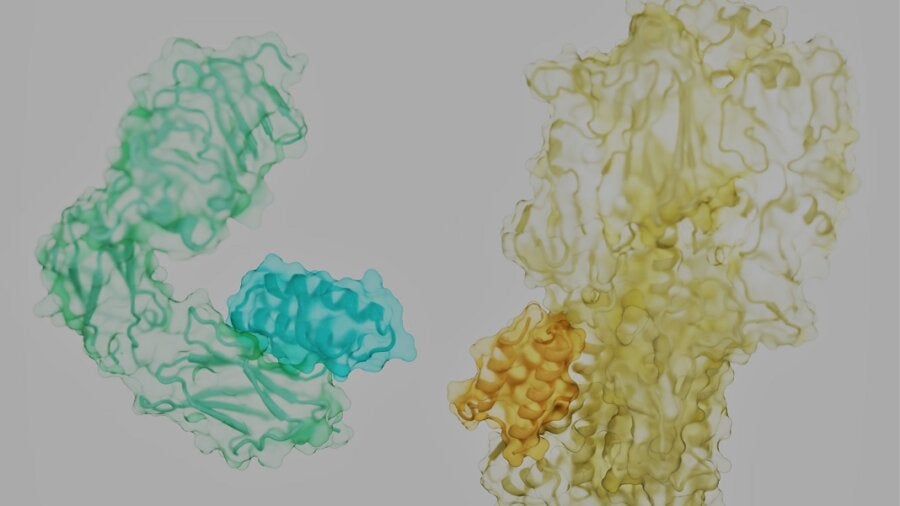
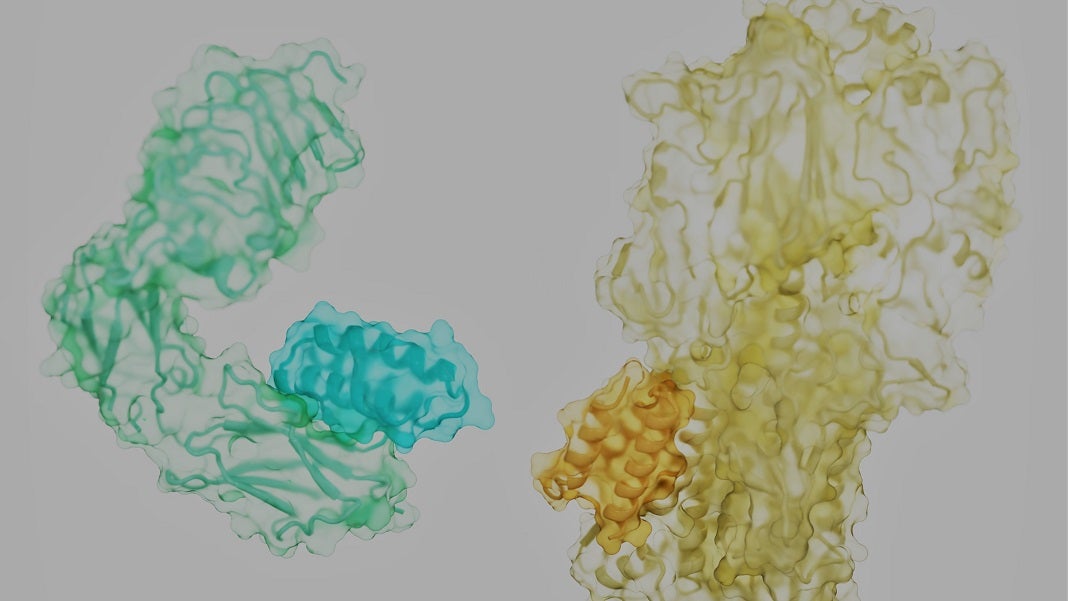
The proteins that management our lives are like rolling tumbleweeds. Every has a tangled, distinctive form, with spiky side-branches dotting its floor. Hidden within the nooks and crannies are the locks to battle our most infamous foes—most cancers, diabetes, infections, and even ageing—if we are able to discover the correct key.
We simply received a common key maker. In a examine revealed at present in Nature, a staff led by Dr. David Baker from the College of Washington developed an algorithm to design tiny protein keys that unlock these targets from scratch. Removed from an ivory tower pursuit, the algorithm tackled probably the most head-scratching drug discovery challenges of our instances: can we design medicine primarily based on the construction of a protein’s lock alone?
They’re not speaking about simply any drug. Somewhat than specializing in small molecules, reminiscent of Tylenol, the staff turned their consideration to protein-like molecules, dubbed “binders.” Whereas they could sound unique, you understand them. Monoclonal antibodies are one instance, which have been key for treating extreme Covid-19 instances. They’re additionally a few of our greatest weapons in opposition to most cancers. However these therapeutic giants wrestle to tunnel into cells, are tough to fabricate, and are sometimes prohibitively costly for widespread use.
What about an alternate? Can we faucet into the facility of contemporary computation and design related however smaller and less complicated medicine which might be simply as—if no more—efficient?
Based mostly on the Baker staff’s examine, the reply is sure. Screening practically half one million candidate binder buildings for 12 protein targets, the algorithm aced its activity, utilizing minimal computational energy in comparison with earlier makes an attempt and highlighting potential hits. It additionally discovered a “cheat code” that made binders extra environment friendly at grabbing onto their targets.
Right here’s the kicker: in contrast to earlier instruments, the software program solely wanted the construction of the goal protein to engineer binder “keys” from scratch. It’s a far less complicated method in comparison with earlier makes an attempt. And since proteins run our inner organic universe, it means the brand new software program key makers might help us unlock the secrets and techniques of our cells’ molecular lives—and intervene once they go awry.
“The flexibility to generate new proteins that bind tightly and particularly to any molecular goal that you really want is a paradigm shift in drug improvement and molecular biology extra broadly,” stated Baker.
Protein Binder What?
Our our bodies are ruled by an enormous consortium of proteins. Like courtesans in a ballroom, every protein bounces across the cell, briefly grabbing onto one other protein earlier than leaving them to seek out the following. Particular pairings can launch mobile plots to set off—or inhibit—dramatic mobile processes. Some might direct a cell to develop or to peacefully cross away. Others might flip a cell cancerous or senescent, leaking poisonous chemical compounds and endangering close by cells.
In different phrases, protein pairings are important for all times. They’re additionally a robust hack for drugs: if any pair triggers a signaling cascade that injures a cell or tissue, we are able to engineer a “doorstop” molecule to actually break up the pairing and cease the illness.
The issue? Think about attempting to separate two intertwined tumbleweeds rolling down a freeway by throwing a brief however versatile stick at them. It appears an not possible activity. However the brand new examine laid out a recipe for achievement: the secret is discovering the place to pry the 2 aside.
Up the Wall
Proteins are sometimes described as beads on chains which might be crumpled into refined 3D buildings. That’s not completely appropriate. The molecular “beads” that make up proteins are extra like humanoid robots, with a stiff trunk and floppy limbs referred to as “facet chains.”
As a protein assembles, it hyperlinks the trunk parts of its constituent amino acids right into a strong spine. Like a fuzzy ball of yarn, the frizz—uncovered facet chains—cowl the protein’s floor. Relying on their place and the spine, they kind pockets {that a} pure protein associate, or a mimic, can readily seize onto.
Earlier research tapped into these pockets to design mimic binders. However the course of is computationally hefty and sometimes depends on identified protein buildings—a beneficial useful resource not all the time out there. One other method is to search out “scorching spots” on a goal protein, however these aren’t all the time accessible to binders.
Right here, the staff took a stab on the downside in a means that’s analogous to rock climbers attempting to scale a brand new wall. The climbers are the binders, the wall is the goal protein floor. Wanting up, there are many handholds and footholds fabricated from facet chains and protein pockets. However the largest ones, the “scorching spots,” can’t essentially maintain the climber for the route.
One other method, the staff defined, is to map out all of the holds, even when some appear weak. This opens a brand new universe of potential binding spots—most will fail, however some combos might surprisingly succeed. A subset of those factors are then challenged with hundreds of climbers, every attempting to determine a promising route. As soon as the highest routes emerge, a second spherical of climbers will discover these routes intimately.
“Following this analogy, we devised a multi-step method to beat” earlier challenges, the staff stated.
To start out, the staff scanned a library of potential protein backbones and an enormous set of sidechain positions that may latch onto a protein goal.
The preliminary pattern sizes have been huge. 1000’s of potential protein spine “trunks” and practically one billion doable sidechain “arms” emerged for each goal.
With the assistance of Rosetta, the protein construction and performance mapping program that Baker’s staff developed, the staff narrowed down the choice to a handful of promising binders.
The choice of these binders depends on “conventional physics” with out tapping into machine studying or deep studying powers, stated Dr. Lance Stewart, chief technique and operations officer on the Institute for Protein Design, the place Baker’s lab is predicated. It “makes this breakthrough much more spectacular.”
Guiding Life
The following massive query: so the binders can bind in silico. However do they really work in cells?
In a proof of idea, the staff picked 12 proteins to check out their algorithm. Amongst these have been proteins carefully concerned in most cancers, insulin, and ageing. One other group zoomed in on battling pathogens, together with floor proteins on the flu or SARS-CoV-2, the virus behind Covid-19.
The staff screened 15,000–100,000 binders for every of the protein targets, and examined high candidates in E. coli micro organism. As anticipated, the binders have been extremely environment friendly at blocking their targets. Some reduce off progress indicators that may result in most cancers. Others focused a standard area of influenza—the flu—that in idea may neutralize a number of strains, paving the way in which for a common flu vaccine. Not even SARS-CoV-2 spared, with the “ultrapotent” binders offering safety in opposition to its invasion in mice (these outcomes have been beforehand revealed).
The examine confirmed that it’s doable to design protein-like medicine from the bottom up. All it takes is the construction of the goal protein.
“The probabilities for utility appear countless,” remarked Dr. Sjors Scheres, joint head of structural research on the MRC Laboratory of Molecular Biology in Cambridge, UK, on Twitter, who wasn’t concerned within the examine.
The algorithm, although highly effective, isn’t excellent. Regardless of discovering tens of millions of potential binders, solely a small fraction of the designs really latched onto their goal. Even the most effective candidates wanted a number of adjustments to their amino acid make-up for optimum binding to a goal.
Nevertheless it’s groundbreaking work for a discipline that might basically change drugs. For now, the tactic and huge dataset “offers a place to begin” to determine how proteins work together inside our cells. These knowledge, in flip, may information even higher computational fashions in a virtuous circle, particularly with an added dose of deep studying magic.
It’ll “additional enhance the velocity and accuracy of design,” stated Stewart. It’s “work that’s already ongoing in our labs.”
Picture Credit score: Longxing Cao, Brian Coventry, David Baker, UW Medication
In search of methods to remain forward of the tempo of change? Rethink what’s doable. Be a part of a extremely curated, unique cohort of 80 executives for Singularity’s flagship Government Program (EP), a five-day, totally immersive management transformation program that disrupts current methods of pondering. Uncover a brand new mindset, toolset and community of fellow futurists dedicated to discovering options to the quick tempo of change on the planet. Click on right here to study extra and apply at present!
[ad_2]